Sample Results for Several Diseases
Here we provide an overview of Sherlock results for several diseases. The tables below list the gene implicated, its LBF, its p-value, whether or not the gene was already implicated in the disease through GWAS (i.e. through significant loci proximal to the gene), and the p-value obtained using our method and a replicate GWAS (when available).
Crohn's Disease
Crohn's is an autoimmune disease of the intestines and has numerous loci identified via GWAS and meta-GWAS. Here we use lymphoblastoid eQTL and a Crohn's meta-GWAS (3,230 cases and 4,829 controls) in Sherlock to search for additional genes. As indicated in the table, many of the resulting genes exhibit strong support in the literature but cannot be identified using GWAS alone. Examples include EFS, a gene that yields a Crohn's-like phenotype in a mouse knockout. Additionally, the target of EFS, the kinase FYN, is among our very top genes (not shown here). Both of these genes are implicated exclusively through trans loci with only modest individual p-values in the GWAS.
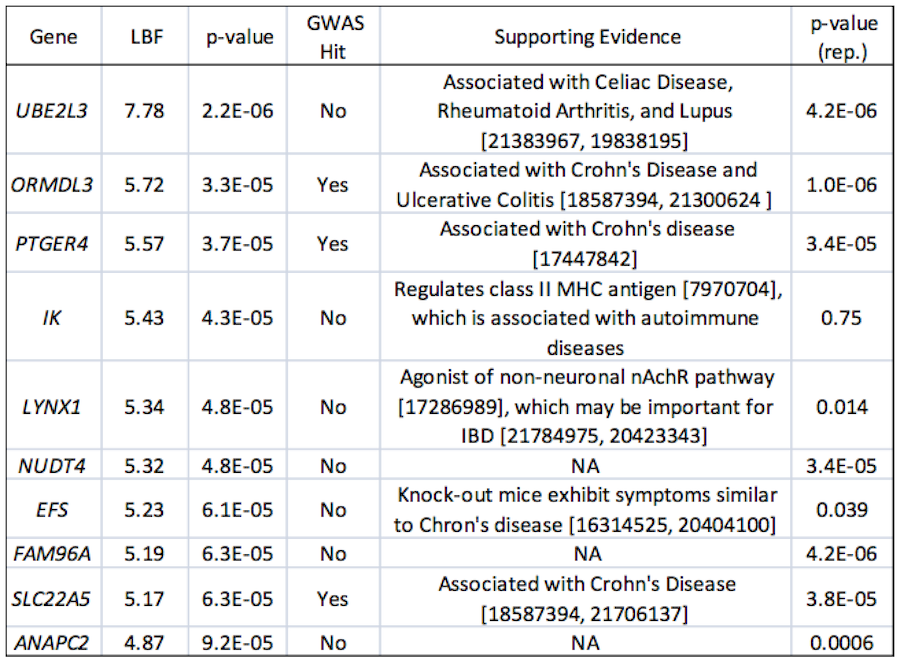 |
Type 2 Diabetes
The above example showcases the use of immune-related eQTL to mine the GWAS of an immune-related disease. Here we intersect eQTL and GWAS for diseases and tissues with a less-obvious connection to the immune system. Using the GWAS of the DIAGRAM consortium (4,549 cases and 5,579 controls) for T2D with liver eQTL yields several genes of modest statistical significance, shown below.
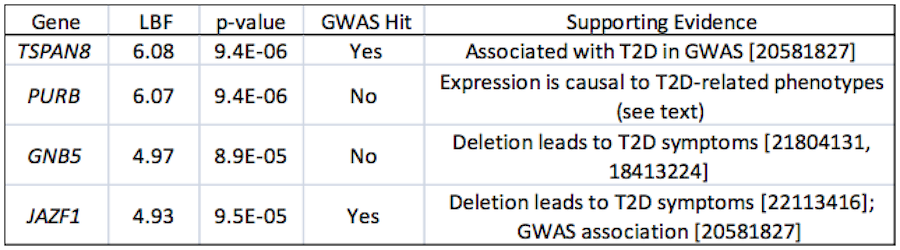 |